Hi, I'm Fang WU!
 Welcome to my personal web page! I am a Ph.D. student at Stanford Computer Science, advised by Yejin Choi.I have great fortune to work with Jure Leskovec, James Zou and Brian Hie during the first-year rotation. I was also a member of the reading group organized by Brian Trippe. Previously, I was a research engineer at Tsinghua University advised by Jinbo Xu. I obtained my Master's degree at Columbia University, advised by Dragomir Radev. It is a profound loss for me to lose Prof. Radev on March 29, 2023 (in memoriam). My research focuses on deep learning algorithms for scientific problems — in particular, LLMs, deep generative models, and geometric deep learning.
Welcome to my personal web page! I am a Ph.D. student at Stanford Computer Science, advised by Yejin Choi.I have great fortune to work with Jure Leskovec, James Zou and Brian Hie during the first-year rotation. I was also a member of the reading group organized by Brian Trippe. Previously, I was a research engineer at Tsinghua University advised by Jinbo Xu. I obtained my Master's degree at Columbia University, advised by Dragomir Radev. It is a profound loss for me to lose Prof. Radev on March 29, 2023 (in memoriam). My research focuses on deep learning algorithms for scientific problems — in particular, LLMs, deep generative models, and geometric deep learning.
Email: fangwu97 [at] stanford [dot] edu
Address: Stanford, CA, USA
Last update time: 2024.06
News and Highlights
![]() [2025/05] One papers is accepted by KDD 2025.
[2025/05] One papers is accepted by KDD 2025.
![]() [2025/05] Two papers are accepted by ACL 2025.
[2025/05] Two papers are accepted by ACL 2025.
![]() [2025/04] One collaborated paper is accepted by ICML 2025. Congrats to Arthor Deng!
[2025/04] One collaborated paper is accepted by ICML 2025. Congrats to Arthor Deng!
![]() [2025/02] One paper on mutant effect prediction is accepted by TMLR.
[2025/02] One paper on mutant effect prediction is accepted by TMLR.
![]() [2024/12] One paper on dynamic surface modeling is accepted by AAAI 2025.
[2024/12] One paper on dynamic surface modeling is accepted by AAAI 2025.
Research Summary
* represents equal contribution and co-first authorship. † denotes the corresponding author(s).
Large Language Models (LLMs)
 Large Language Models are Good Relational Learners.
Large Language Models are Good Relational Learners.
Fang Wu, Vijay Prakash Dwivedi, Jure Leskovec†
ACL 2025
[Paper]
[Code]
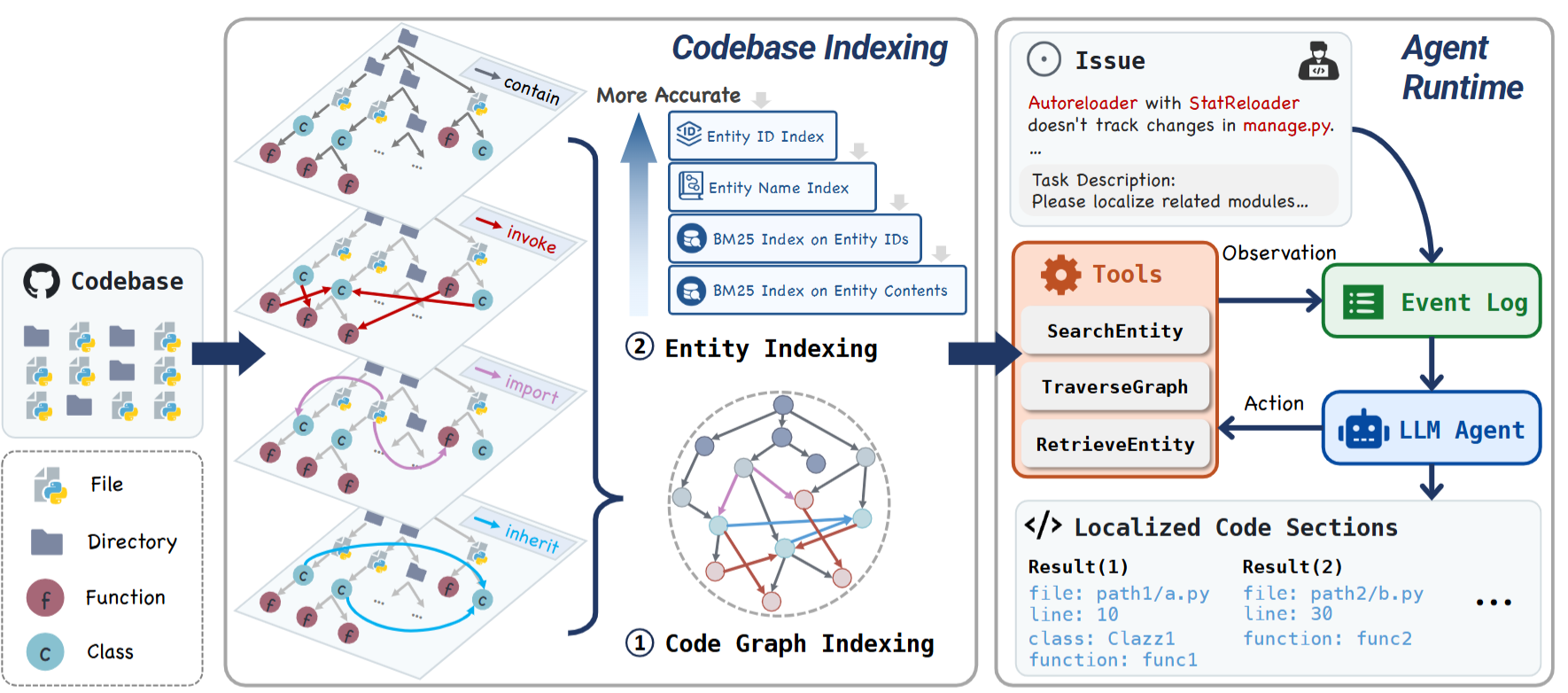 LocAgent: Graph-Guided LLM Agents for Code Localization.
LocAgent: Graph-Guided LLM Agents for Code Localization.
Zhaoling Chen*, Xiangru Tang*, Gangda Deng*, Fang Wu, Jialong Wu, Zhiwei Jiang, Viktor Prasanna, Arman Cohan, Xingyao Wang†
ACL 2025
[Paper]
[Code]
 When to Trust Context: Self-Reflective Debates for Context Reliability
When to Trust Context: Self-Reflective Debates for Context Reliability
Zeqi Zhou*, Fang Wu*†, Shayan Talaei*, Haokai Zhao, Cheng Meixin, Tinson Xu, Amin Saberi, Yejin Choi
Under Review
[Paper]
[Code]
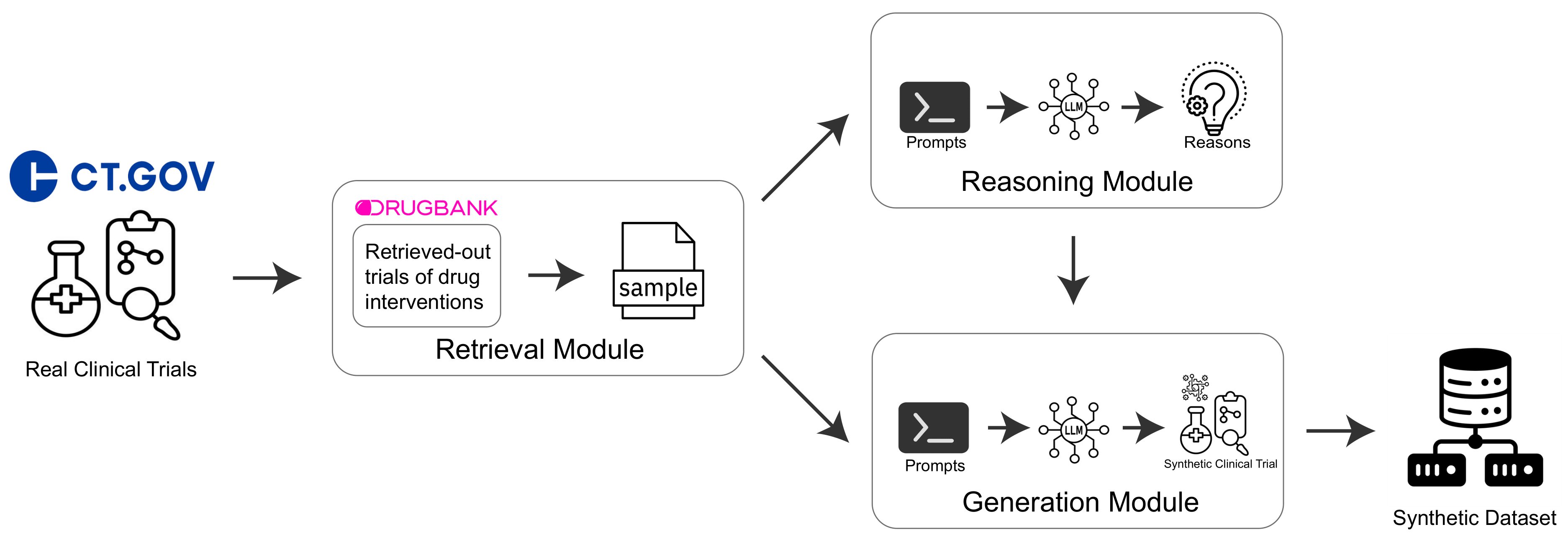 Retrieval-Reasoning Large Language Model-based Synthetic Clinical Trial Generation
Retrieval-Reasoning Large Language Model-based Synthetic Clinical Trial Generation
Zerui Xu, Fang Wu, Tianfan Fu, Yue Zhao†
Under Review
[Paper]
[Code]
LLMs for Science
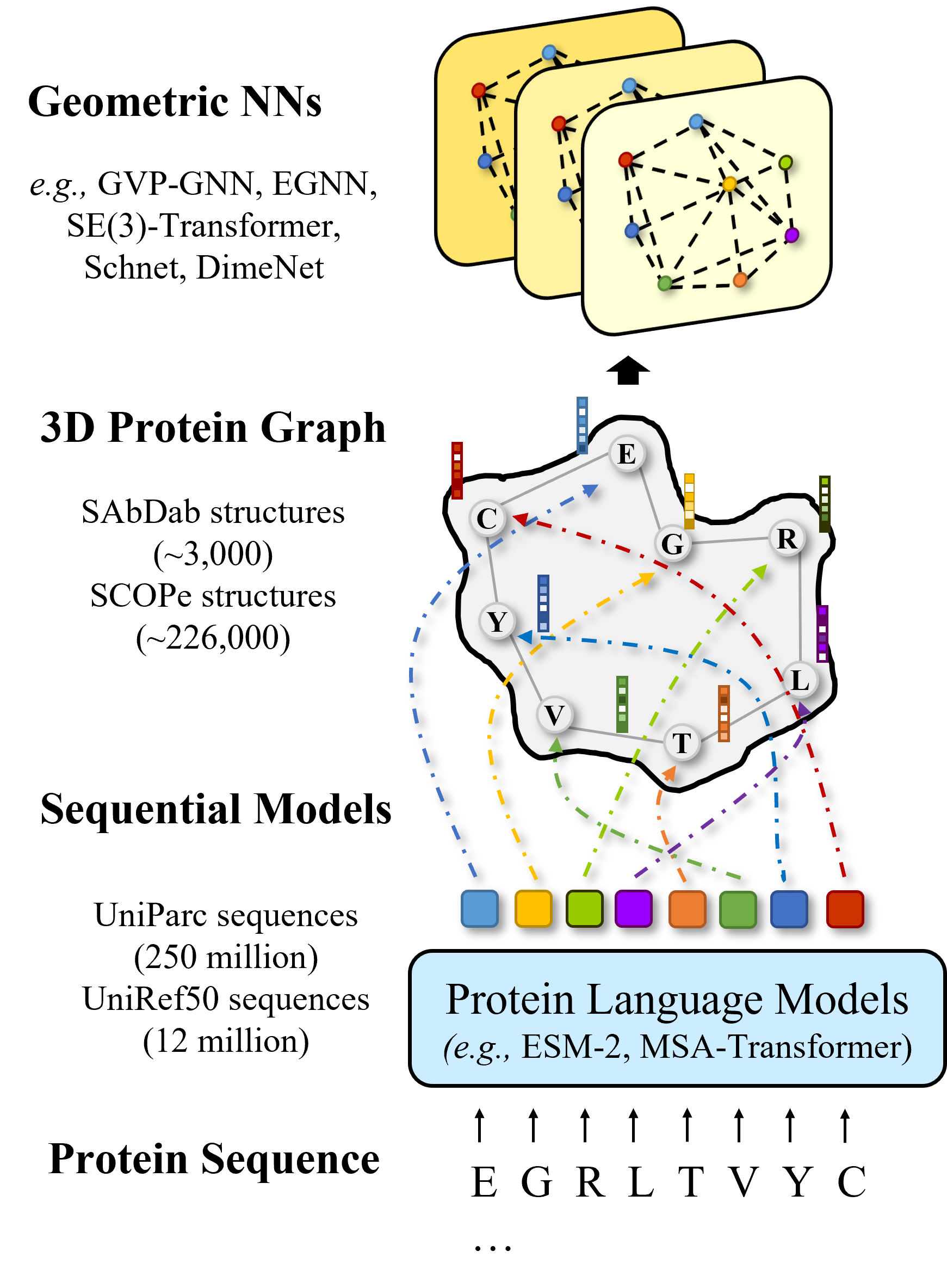 Integration of Pre-trained Protein Language Mdels into Geometric Deep Learning Networks.
Integration of Pre-trained Protein Language Mdels into Geometric Deep Learning Networks.
Fang Wu, Liong Wu, Dragomir Radev, Jinbo Xu, Stan Z. Li†
Communications Biology (2023)
[Paper]
[Code]
Graph Neural Networks (GNNs)
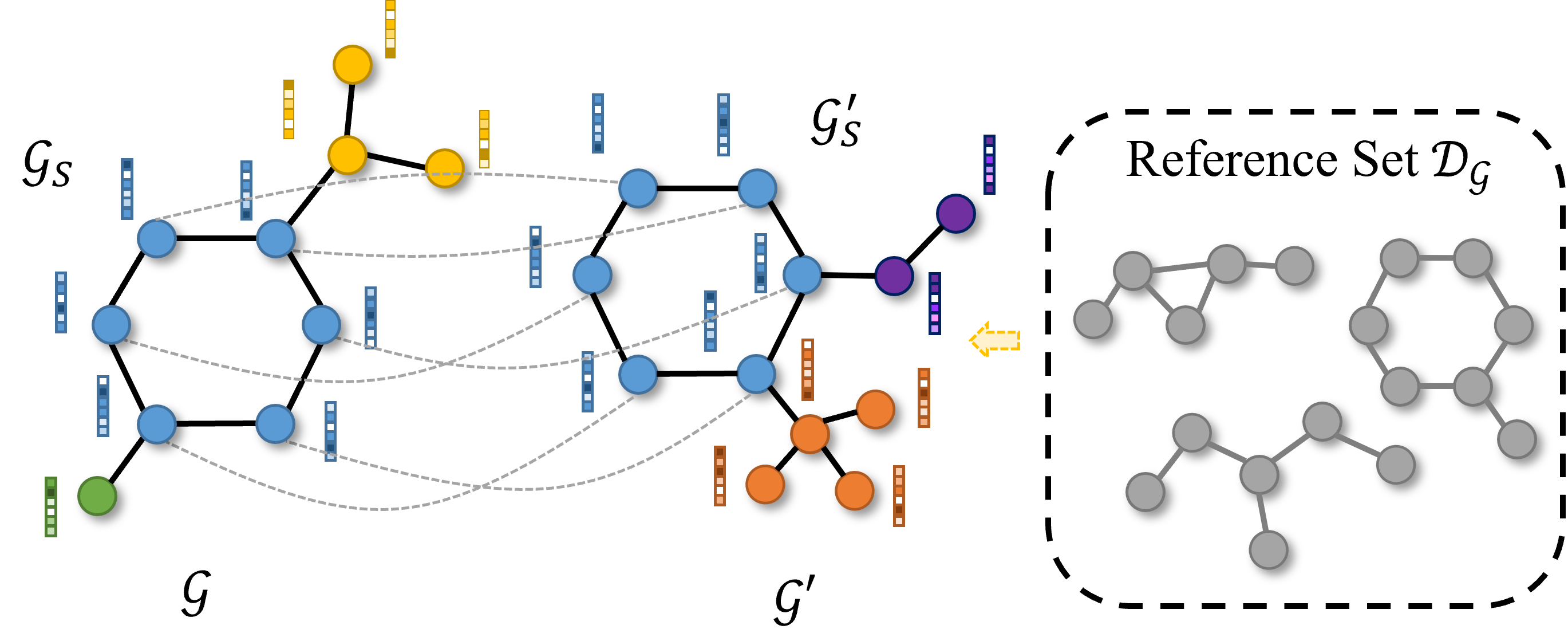 Rethinking Explaining Graph Neural Networks via Non-parametric Subgraph Matching
Rethinking Explaining Graph Neural Networks via Non-parametric Subgraph Matching
Fang Wu, Siyuan Li, Dragomir Radev, Stan Z. Li†
ICML 2023
[Paper]
[Code]
 Discovering and Explaining the Representation Bottleneck of Graph Neural Networks from Multi-order Interactions
Discovering and Explaining the Representation Bottleneck of Graph Neural Networks from Multi-order Interactions
Fang Wu*, Siyuan Li*, Dragomir Radev, Stan Z. Li†
IEEE TKDE (2024)
[Paper]
[Code]
 InsertGNN: A Hierarchical Graph Neural Network for the TOEFL Sentence Insertion Problem
InsertGNN: A Hierarchical Graph Neural Network for the TOEFL Sentence Insertion Problem
Fang Wu†, Stan Z. Li
EMNLP 2024 Findings
[Paper]
[Data]
Deep Generative Models for Drug Design
 D-Flow: Multi-modality Flow Matching for D-peptide Design.
D-Flow: Multi-modality Flow Matching for D-peptide Design.
Fang Wu*, Tinson Xu*, Shuting Jin*, Xiangru Tang, Zerui Xu, James Zou, Brian Hie†
Under review
[Paper]
[Code]
 SurfDesign: Effective Protein Design on Molecular Surfaces.
SurfDesign: Effective Protein Design on Molecular Surfaces.
Fang Wu, Shuting Jin, Jianmin Wang, Zerui Xu, xiangxiang Zeng, Jinbo Xu†
Under review
[Paper]
[Code]
 BC-Design: A Biochemistry-Aware Framework for High-Precision Inverse Protein Folding.
BC-Design: A Biochemistry-Aware Framework for High-Precision Inverse Protein Folding.
Xiangru Tang*, Xinwu Ye*, Fang Wu*, Yanjun Shao, Yin Fang, Siming Chen, Dong Xu, Mark Gerstein†
Under review
[Paper]
[Code]
 Surface-based Peptide Design with Multi-modal Flow Matching.
Surface-based Peptide Design with Multi-modal Flow Matching.
Fang Wu*, Shuting Jin, Zhengyuan Zhou, Xiangxiang Zeng, Jure Leskovec, Jinbo Xu†
KDD 2025
[Paper]
 A Survey of Generative AI for de novo Drug Design: New Frontiers in Molecule and Protein Generation.
A Survey of Generative AI for de novo Drug Design: New Frontiers in Molecule and Protein Generation.
Xiangru Tang*, Howard Dai*, Elizabeth Knight*, Fang Wu,, Yunyang Li, Tianxiao Li, Mark Gerstein†
Briefings in Bioinformatics (2024)
[Paper]
[Github Repo.]
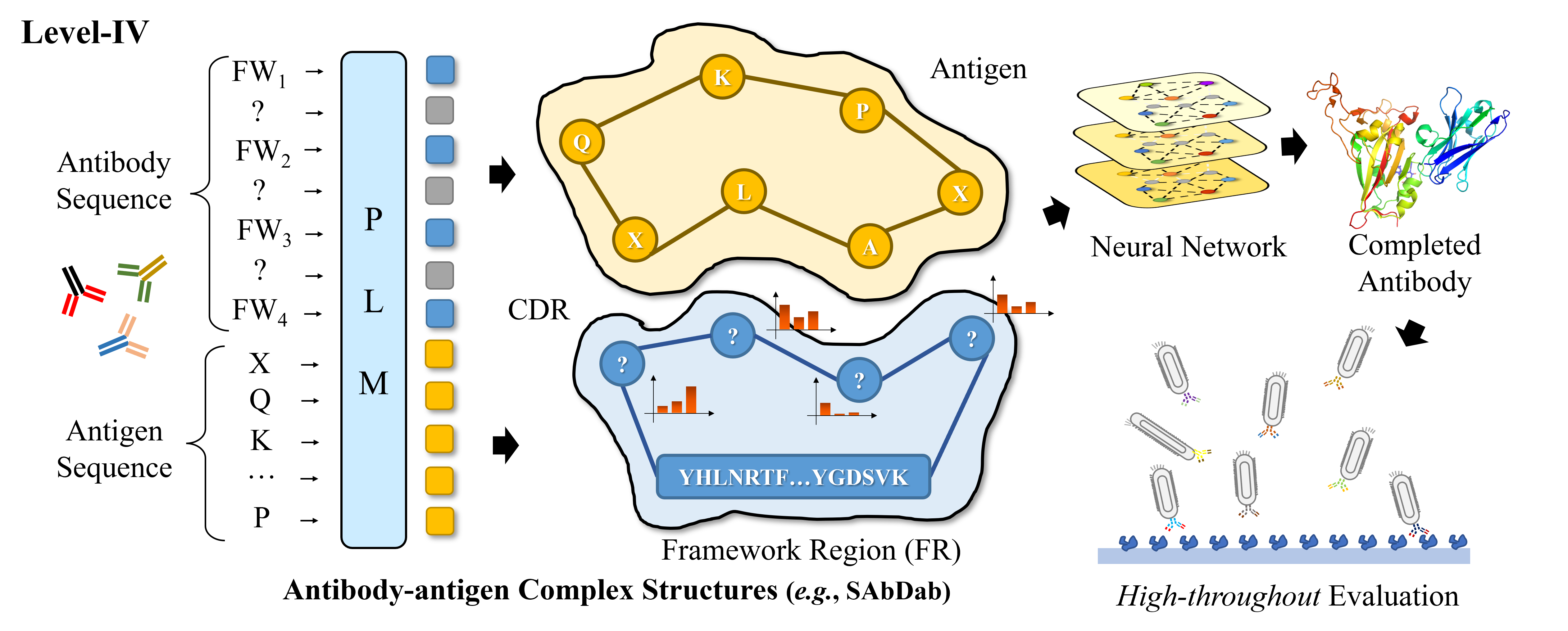 A Hierarchical Training Paradigm for Antibody Structure-sequence Co-design
A Hierarchical Training Paradigm for Antibody Structure-sequence Co-design
Fang Wu, Stan Z. Li†
NeurIPS 2023
[Paper]
3D Geometric Deep Learning
 PoseX: AI Defeats Physics Approaches on Protein-Ligand Cross Docking.
PoseX: AI Defeats Physics Approaches on Protein-Ligand Cross Docking.
Yize Jiang*, Xinze Li*, Yuanyuan Zhang*, Jin Han*, Youjun Xu*, Ayush Pandit, Zaixi Zhang, Mengdi Wang, Mengyang Wang, Chong Liu, Guang Yang, Yejin Choi, Wu‑Jun Li†, Tianfan Fu†, Fang Wu†, Junhong Liu†
Under Review
[Paper]
[Code]
[Webpage]
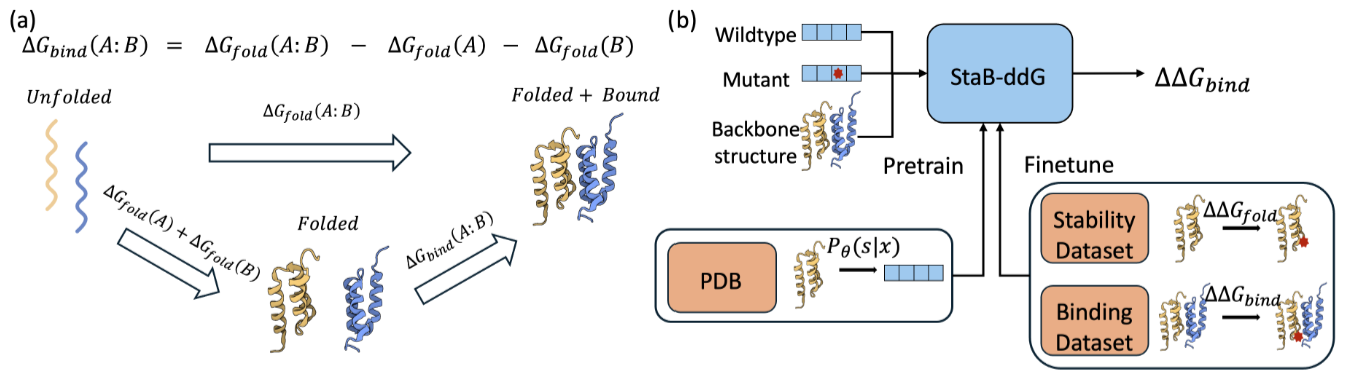 StaB-ddG: Predicting mutational effects on protein binding from folding energy.
StaB-ddG: Predicting mutational effects on protein binding from folding energy.
Arthur Deng, Karsten D. Householder, Fang Wu, Sebastian Thrun, K. Christopher Garcia, Brian L. Trippe†
ICML 2025
[Paper]
[Code]
 Dynamics-inspired Structure Hallucination for Protein-protein Interaction Modeling.
Dynamics-inspired Structure Hallucination for Protein-protein Interaction Modeling.
Fang Wu, Stan Z. Li†
TMLR (2025)
[Paper]
[Code]
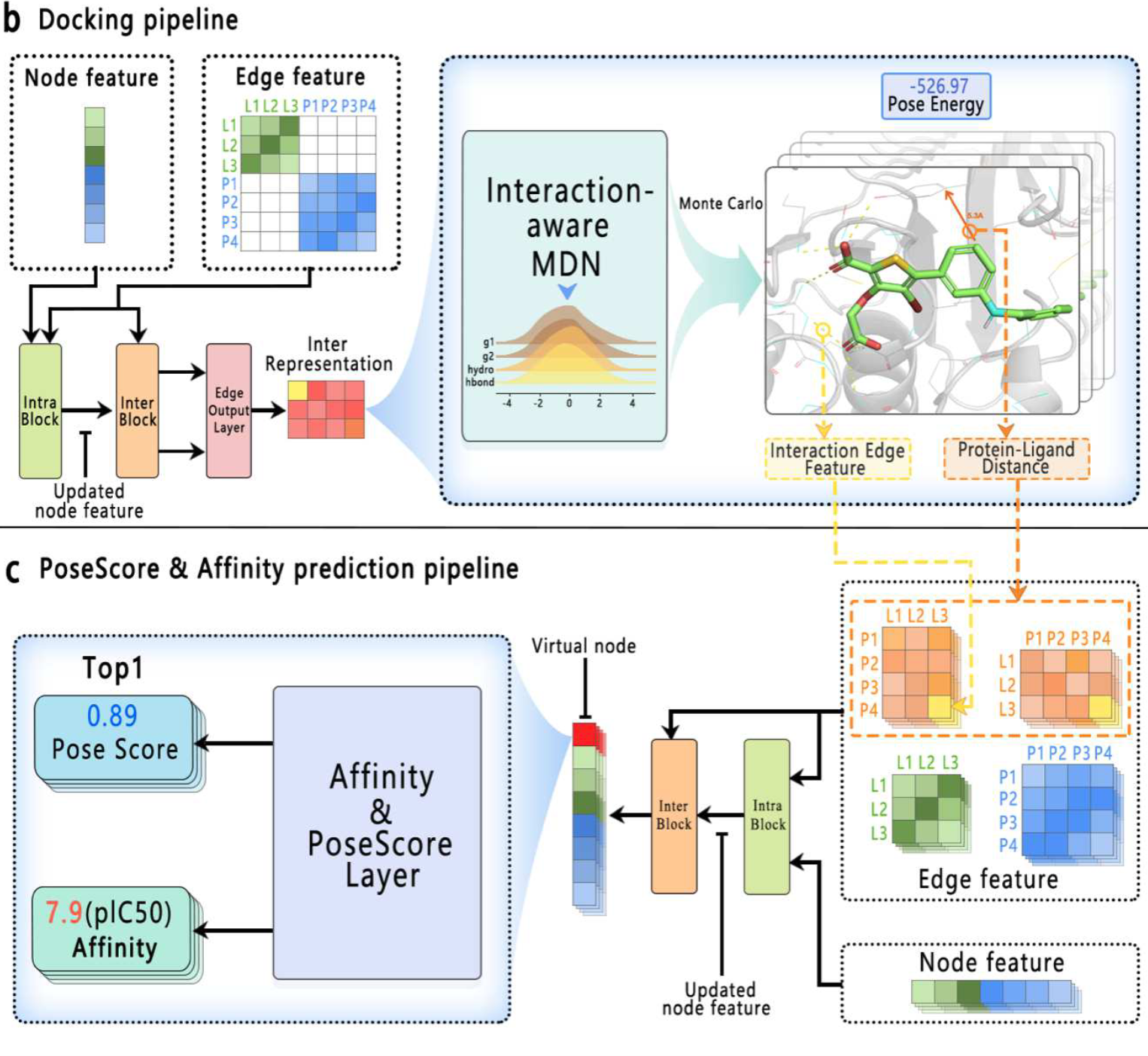 Interformer: An Interaction-Aware Model for Protein-Ligand Docking and Affinity Prediction.
Interformer: An Interaction-Aware Model for Protein-Ligand Docking and Affinity Prediction.
Houtim Lai†, Longyue Wang†, Ruiyuan Qian, Juhong Huang, Peng Zhou, Geyan Ye, Fandi Wu, Fang Wu, Xiangxiang Zeng, Wei Liu
Nature Communications (2024)
[Paper]
[Code]
 Surface-VQMAE: Vector-quantized Masked Auto-encoders on Molecular Surfaces.
Surface-VQMAE: Vector-quantized Masked Auto-encoders on Molecular Surfaces.
Fang Wu, Stan Z. Li†
ICML 2024
[Paper]
[Code]
 Molformer: Motif-based Transformer on 3D Heterogeneous Molecular Graphs.
Molformer: Motif-based Transformer on 3D Heterogeneous Molecular Graphs.
Fang Wu, Dragomir Radev, Stan Z. Li†
AAAI 2023
[Paper]
[Code]
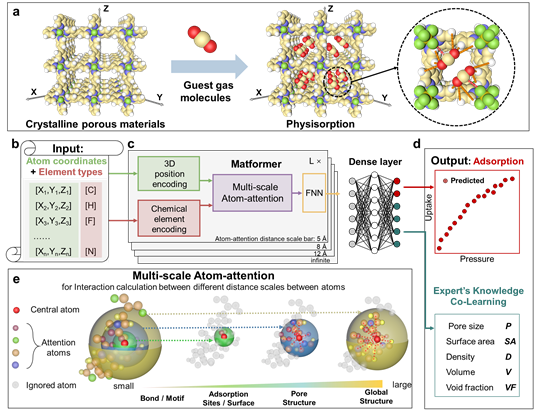 Direct Prediction of Gas Adsorption via Spatial Atom Interaction Learning.
Direct Prediction of Gas Adsorption via Spatial Atom Interaction Learning.
Jiyu Cui*, Fang Wu*, Wen Zhang*, Lifeng Yang*, Jianbo Hu, Yin Fang, Peng Ye, Qiang Zhang, Xian Suo, Yiming Mo, Xili Cui, Huajun Chen†, Huabin Xing†
Nature Communications (2023)
[Paper]
[Code]
Molecular Property Prediction
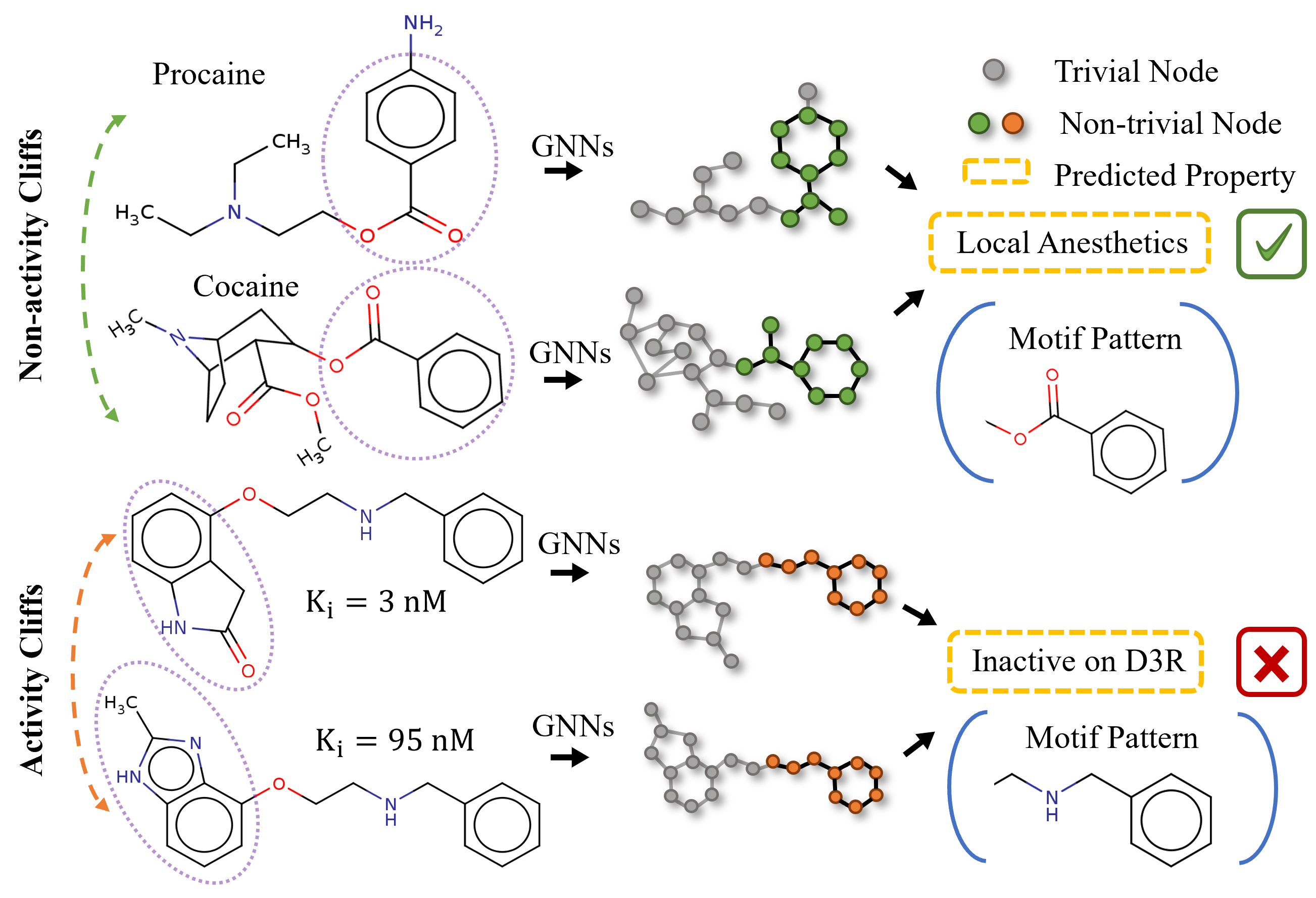 A Semi-supervised Molecular Learning Framework for Activity Cliff Estimation.
A Semi-supervised Molecular Learning Framework for Activity Cliff Estimation.
Fang Wu†
IJCAI 2024
[Paper]
[Code]
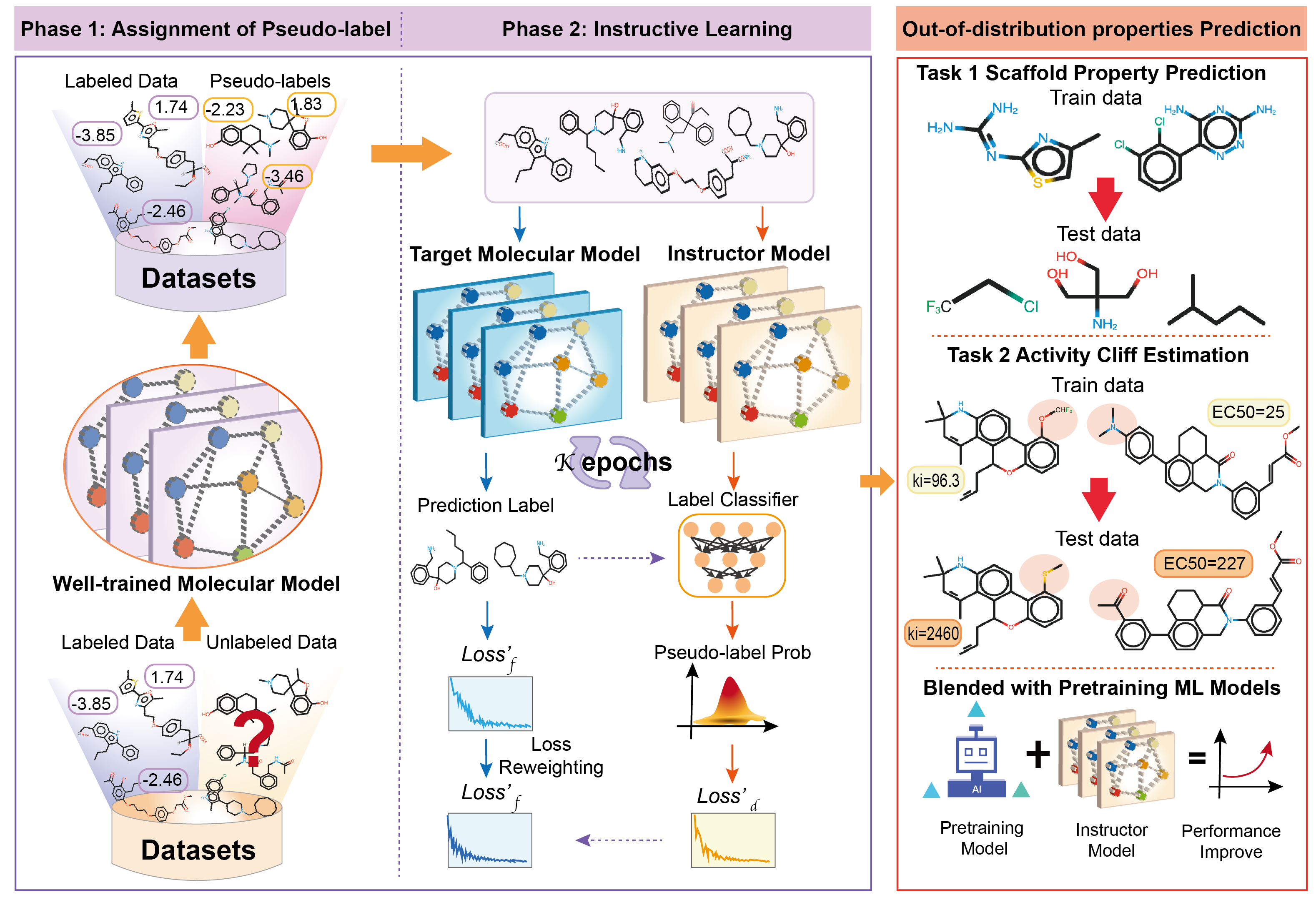 A Semi-supervised Molecular Learning Framework for Activity Cliff Estimation.
A Semi-supervised Molecular Learning Framework for Activity Cliff Estimation.
Fang Wu*†, Shuting Jin*, Siyuan Li, Stan Z. Li
NeurIPS 2024
[Paper]
[Code]
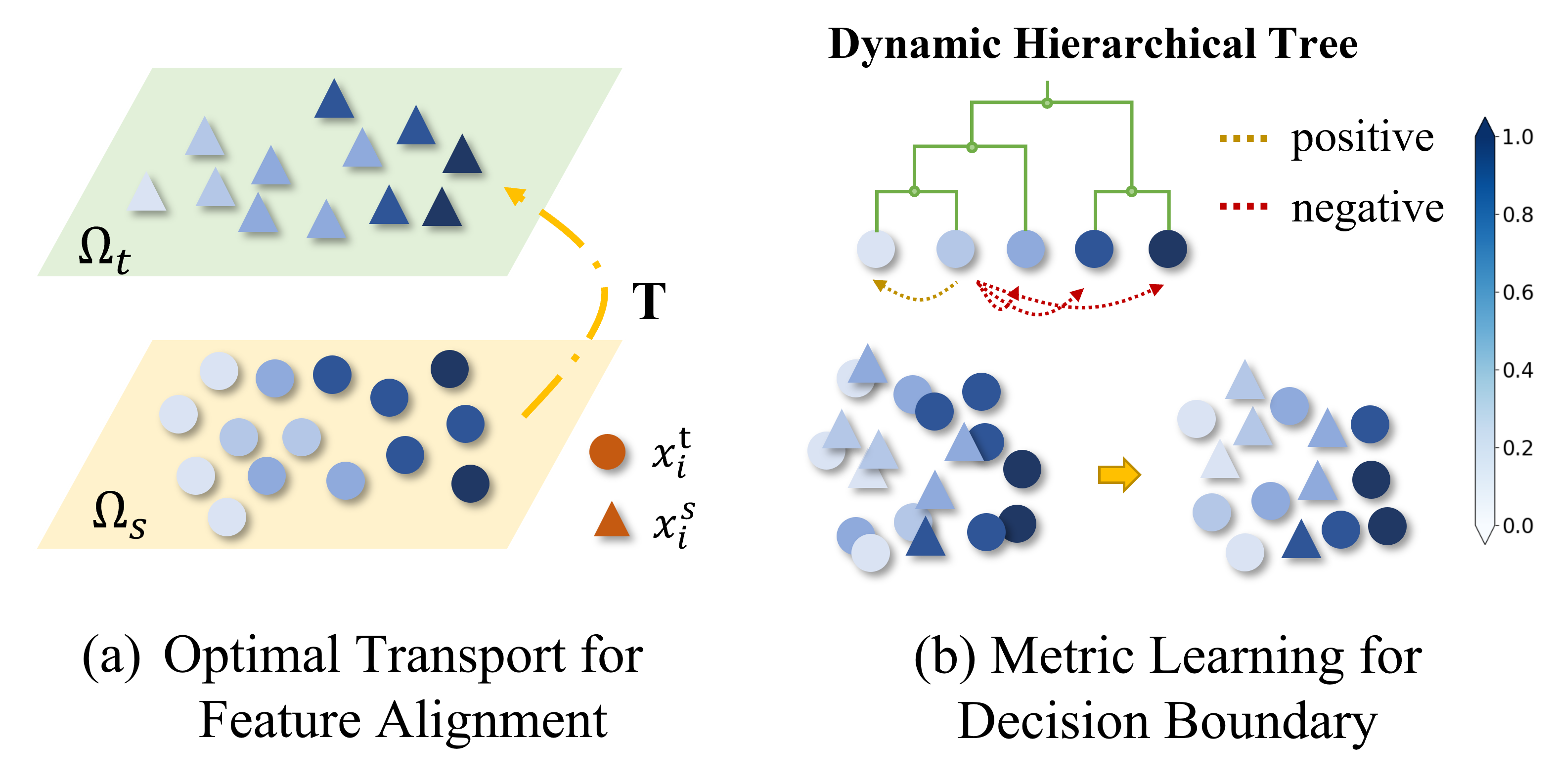 Metric Learning-enhanced Optimal Transport for Biochemical Regression Domain Adaptation
Metric Learning-enhanced Optimal Transport for Biochemical Regression Domain Adaptation
Fang Wu*, Nicolas Courty*, Shuting Jin*, Stan Z. Li†
Patterns (2023)
[Paper]
[Code]
Molecular Dynamics (MD) Simulations
 Generalized Implicit Neural Representations for Dynamic Molecular Surface Modeling.
Generalized Implicit Neural Representations for Dynamic Molecular Surface Modeling.
Fang Wu, Bozhen Hu, Stan Z. Li†
AAAI 2025
[Paper]
 DiffMD: A Geometric Diffusion Model for Molecular Dynamics Simulations
DiffMD: A Geometric Diffusion Model for Molecular Dynamics Simulations
Fang Wu, Stan Z. Li†
AAAI 2023 (Oral)
[Paper]
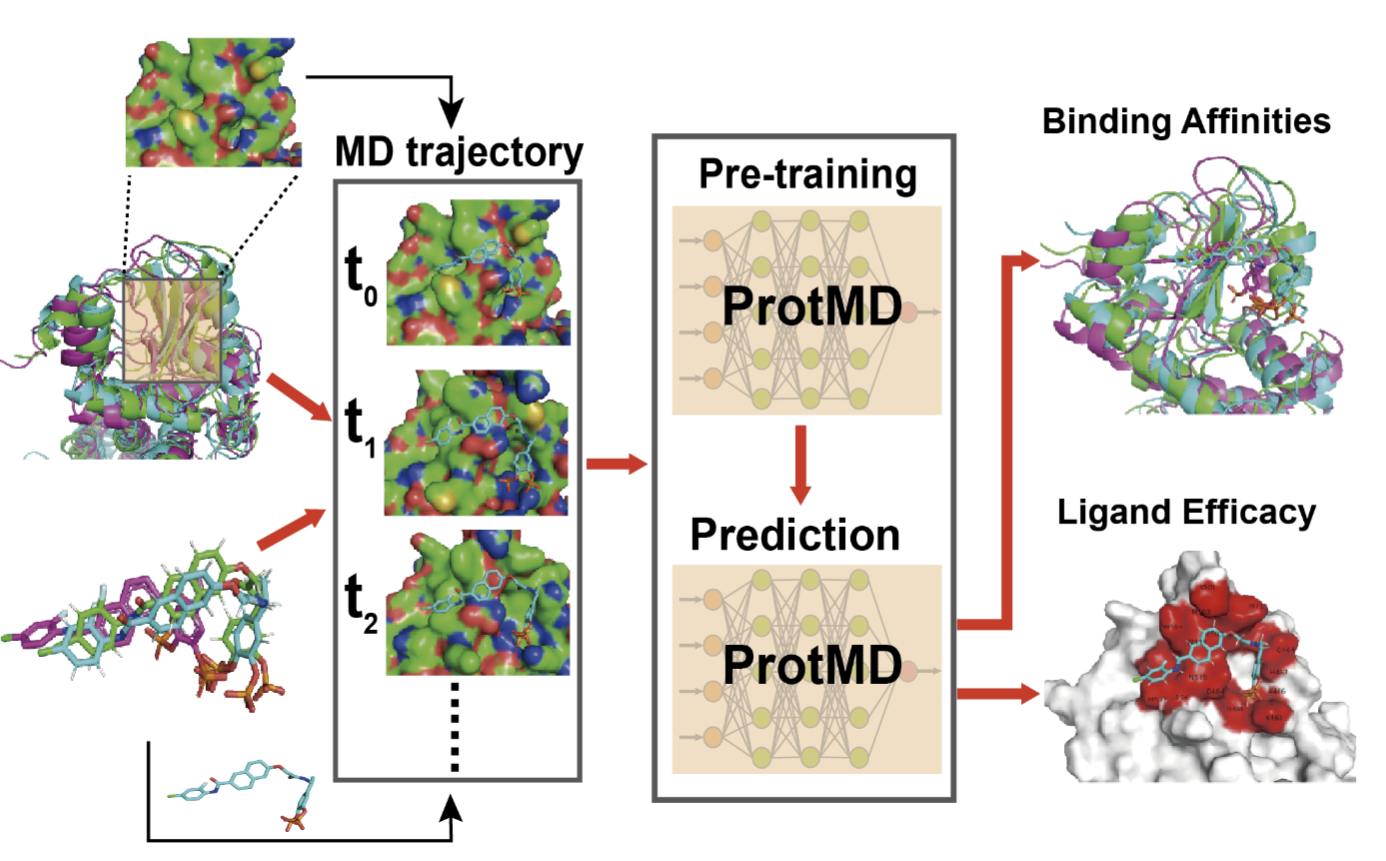 Pretraining of Equivariant Graph Matching Networks with Conformation Flexibility for Drug Binding
Pretraining of Equivariant Graph Matching Networks with Conformation Flexibility for Drug Binding
Fang Wu*, Shuting Jin*, Yinghui Jiang*, Xurui Jin, Bowen Tang, Zhangming Niu, Qiang Zhang, Xiangxiang Zeng, Stan Z. Li†
Advanced Science (2022)
[Paper]
[Code]
Computer Vision
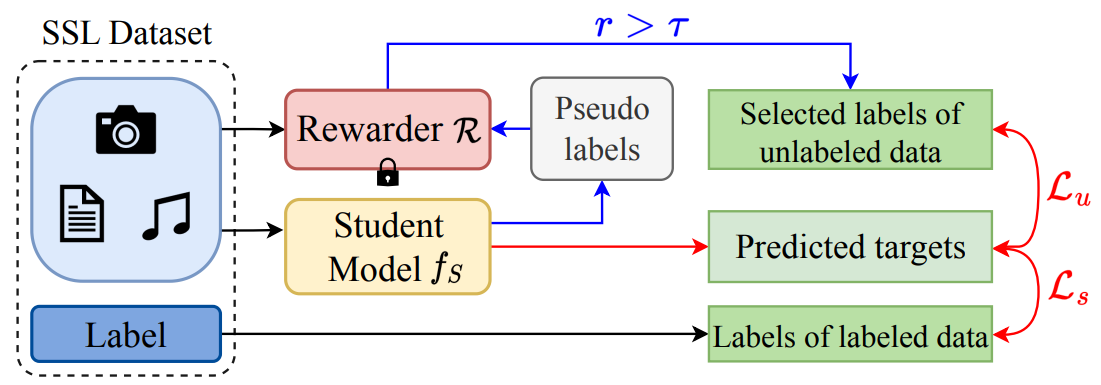 SemiReward: A General Reward Model for Semi-supervised Learning
SemiReward: A General Reward Model for Semi-supervised Learning
Siyuan Li*, Weiyang Jin*, Zedong Wang, Fang Wu,, Zicheng Liu, Cheng Tan, Stan Z. Li†
ICLR 2024
[Paper]
[Code]
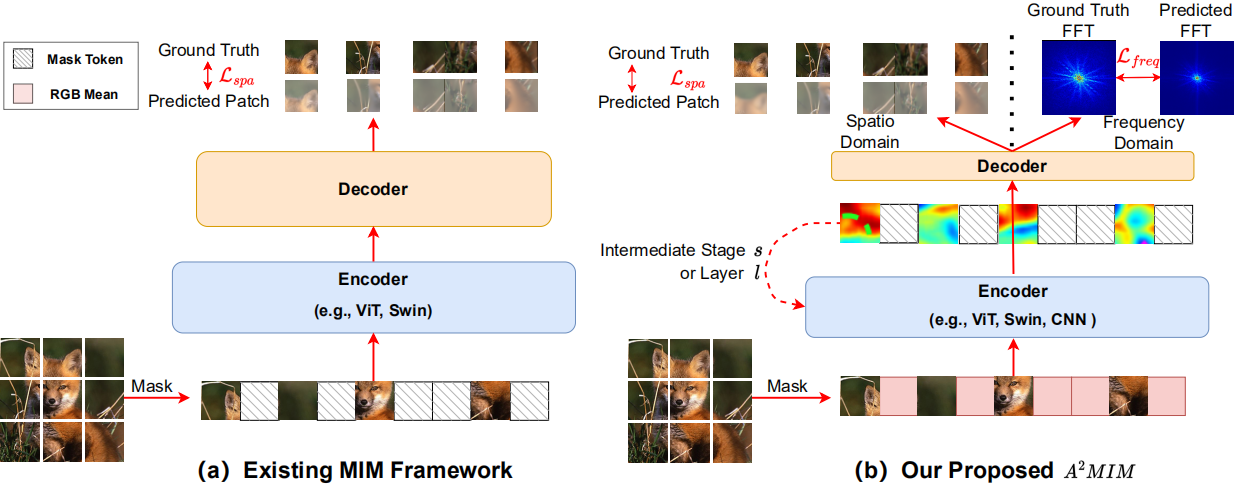 Architecture-Agnostic Masked Image Modeling: From ViT back to CNN
Architecture-Agnostic Masked Image Modeling: From ViT back to CNN
Siyuan Li*, Di Wu*, Fang Wu, Zelin Zang, Kai Wang, Lei Shang, Baigui Sun, Hao Li, Stan Z. Li†
ICML 2023
[Paper]
[Code]
Education
 Stanford University, 2024-now
Stanford University, 2024-now
• Ph.D. in Computer Science
 Columbia University, 2019-2021
Columbia University, 2019-2021
• Master of Science
• GPA: 3.51/4.0
Industry Experience
 Research Scientist (2023.06-2024.06)
Research Scientist (2023.06-2024.06)
• BioMap
• Led by Le Song
 Research Intern (2022.01-2022.07)
Research Intern (2022.01-2022.07)
• MindRank
• Led by Zhangming Niu
Research Experience
Before joining Stanford University, I feel fortunate to be a research assistant/engineer advised by Jinbo Xu at Tsinghua University and Stan Z. Li at Westlake University, and recieved guidance as a visiting student from Huajun Chen, Xiang Bai and Danny Lan.
 Research Student (2024.09-2024.12)
Research Student (2024.09-2024.12)
• Arc Institute
• Advised by Brian Hie
 Research Engineer (2022.08-2023.05)
Research Engineer (2022.08-2023.05)
• Tsinghua University
• Advised by Jinbo Xu
Professional Services
Reviewer: ICLR 2024-2025 , NeurIPS 2023-2025, ICML 2025 , CVPR 2025, ICCV 2025, KDD 2025, AISTATS 2025, IJCAI 2025, ML4H 2023-2024, TMLR, IEEE TNNLS
Teaching: CS224N (2024 Winter)
Acknowledgement
My study cannot be possible without the support from my awesome friends, mentors, and collaborators! Check out some of them:
Prof. Jure Leskovec, Prof. James Zou, Prof. Brian Hie, Prof. Brian Trippe, Dr. Auther Deng at Stanford University.
Prof. Dragomir Radev, Dr. Xiangru Tang at Yale University. R.I.P. to Dr. Dragomir.
Aside from university collaborations, I also collaborated with many industrial AIDD companies, including MindrankAI, MoleculeMind, and Biomap
Dr. Zhangming Niu, Dr. Xurui Jin, and Dr. Yinghui Jiang at MindrankAI.
Dr. Xiaoyang Jing, Dr. Tenglong Wang, Dr. Wuwei Tan at MoleculeMind.

 Central University of Economics and Finance, 2015-2019
Central University of Economics and Finance, 2015-2019  Research Assistant (2021.11-2022.07)
Research Assistant (2021.11-2022.07)  Visiting Student (2021.03-2021.10)
Visiting Student (2021.03-2021.10)